Covid-19 is clearly one of the most significant health crises of modern times, and our nonprofit is trying to contribute to the fight as much as we can. We need your help though! YOU can join the fight alongside us.
Donate your computer to find COVID-19 drugs!
Join Quarantine@Home!
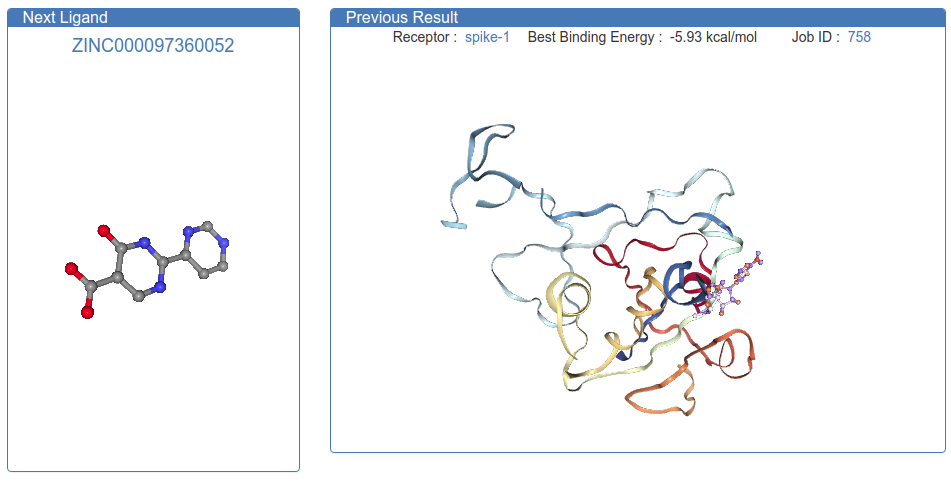
Last month we very quickly build Quarantine@Home, a completely open-source software package that simulates small-molecule drug binding to COVID-19 proteins. We are one of the winners of the Coronavirus Global Hackathon. Since then, we have amassed about 50 volunteers who have collectively simulated ~250,000 (and growing) drugs. You can see the results here. WE NEED MORE VOLUNTEERS. Despite computing 250k calculations already, there are nearly 1-billion possible drugs in the database.
To join : Windows, Macintosh, and Linux clients are all available!
Join Folding@Home!
After we rolled out Quarantine@Home, we found out that Folding@Home very quickly retrofitted their platform to perform “Free Energy Perturbations” (FEP) to simulate drugs bound to coronavirus proteins. This is an impressive upgrade to their network, which primarily has been used to simulate proteins “folding” by themselves. We’re now collaborating with Folding@Home, as their simulations need docked poses to begin with. We are hoping to use the docking algorithms (based on Autodock developed by Scripps Research) to pre-screen from the billion possible drugs, and Folding@Home can perform much more comprehensive simulations of the best binders.